The sequencing service is used in an increasing number of different areas of life. For many years, automated sequencers have come to the rescue of DNA analysis. But how exactly is this process going from behind the scenes? In this article, we will try to explain the technical aspects of the sequencing service in the technology of long DNA readings, i.e. nanopore technology.
Practical nanopore sequencing
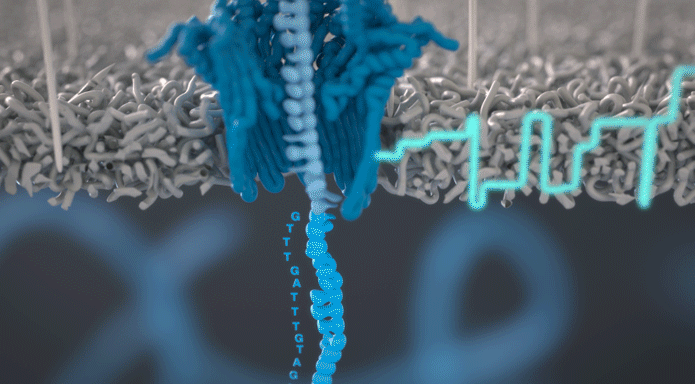
Pores are protein structures that allow ions to flow through the biological membrane. Some of them create channels with a diameter that allows translocation not only of ions but also of large macromolecules. Among the pores with the above properties, one can distinguish, among others alpha-haemolysins, mspA – pores produced by mycobacteria or csgG proteins, forming secretory channels in E. coli.
The pores used in the sequencing process are called nanopores. These are genetically modified proteins that have been previously selected for the efficiency of nucleic acid translocation and distributed in a synthetic membrane. The transition of the nucleic acid from one side of the membrane to the other is accompanied by a change in electric potential, characteristic of a particular nucleotide sequence. We wrote about how an electrical signal is converted into a nucleotide sequence in the publication on the topic of basecalling. For the DNA or RNA strand to move in a controlled manner along the nanopore, it is necessary to attach a protein molecule called an adapter to it. The protein adapter performs two important functions: it unravels the helix and imparts a precisely defined speed at which nucleic acid molecules pass through the pore.
The technology of long DNA readings
As a platform, Nanopore offers readings with N50> 10 kb, thus far superior to the second-generation sequencing technologies. The length of the reading is only limited by the length of the analyzed DNA fragment. Long readings have a wide range of applications in the genomics of model and non-model organisms as well as in clinical diagnostics. First of all, they increase the probability of correct mapping, thanks to which they enable the assembly of de novo genomes, i.e. without prior knowledge of the reference sequence for a given organism. In addition, the analysis of tens of thousands of nucleotides in a single reading provides a unique opportunity to perform the analysis of structural chromosomal aberrations, thus enabling the detection and analysis of extensive structural mutations such as deletions, insertions, inversions and translocations. Another advantage of the 3rd generation technology is the ability to analyze large repeated regions, not available for sequencing based on generating short reads (<1000 bp). The detection and accurate quantification of a variable number of repeated elements are particularly useful in the diagnosis of diseases such as fragile X syndrome and Huntington’s chorea. Additionally, it enables correlating the functionality of the protein with the number of repeated copies of the gene encoding it, as in the case of AMY1. Nanopores also allow for the sequencing of native nucleic acids, thus eliminating errors related to amplification, which may result, for example, from the preferential binding of primers to some DNA templates. The lack of the necessity to perform the PCR reaction also results in the possibility of analyzing the methylation level of the sequenced strands.